rnalysis.filtering.CountFilter.pca
- CountFilter.pca(samples: GroupedColumns | Literal['all'] = 'all', n_components: PositiveInt = 3, power_transform: bool = True, labels: bool = True, title: str | Literal['auto'] = 'auto', title_fontsize: float = 20, label_fontsize: float = 16, tick_fontsize: float = 12, proportional_axes: bool = False, plot_grid: bool = True, legend: List[str] | None = None) Tuple[PCA, List[Figure]]
Performs Principal Component Analysis (PCA), visualizing the principal components that explain the most variance between the different samples. The function will standardize the data prior to PCA, and then plot the requested number of pairwise PCA projections.
- Parameters:
samples ('all' or list.) – A list of the sample names and/or grouped sample names to be plotted. All specified samples must be present in the CountFilter object. To draw multiple replicates of the same condition in the same color, they can be grouped in an inner list. Example input: [[‘SAMPLE1A’, ‘SAMPLE1B’, ‘SAMPLE1C’], [‘SAMPLE2A’, ‘SAMPLE2B’, ‘SAMPLE2C’],’SAMPLE3’ , ‘SAMPLE6’]
n_components (int >=2 (default=3)) – number of Principal Components to plot (minimum is 2). RNAlysis will generate a pair-wise scatter plot between every pair of Principal Components.
labels (bool (default=True)) – if True, RNAlysis will display labels with the sample names next to each sample on the graph.
power_transform (bool (default=True)) – if True, RNAlysis will apply a power transform (Box-Cox) to the data prior to standartization and principal component analysis.
title (str or 'auto' (default='auto')) – The title of the plot. If ‘auto’, a title will be generated automatically.
title_fontsize (float (default=30)) – determines the font size of the graph title.
label_fontsize (float (default=15)) – determines the font size of the X and Y axis labels.
tick_fontsize (float (default=10)) – determines the font size of the X and Y tick labels, and the sample name labels. .
proportional_axes (bool (default=False)) – if True, the dimensions of the PCA plots will be proportional to the percentage of variance explained by each principal component.
plot_grid (bool (default=True)) – if True, will draw a grid on the PCA plot.
legend (list of str, or None (default=None)) – if enabled, display a legend on the PCA plot. Each entry in the ‘legend’ parameter corresponds to one group of samples (one color on the graph), as defined by the parameter ‘samples’
- Returns:
A tuple whose first element is an sklearn.decomposition.pca object, and second element is a list of matplotlib.axis objects.
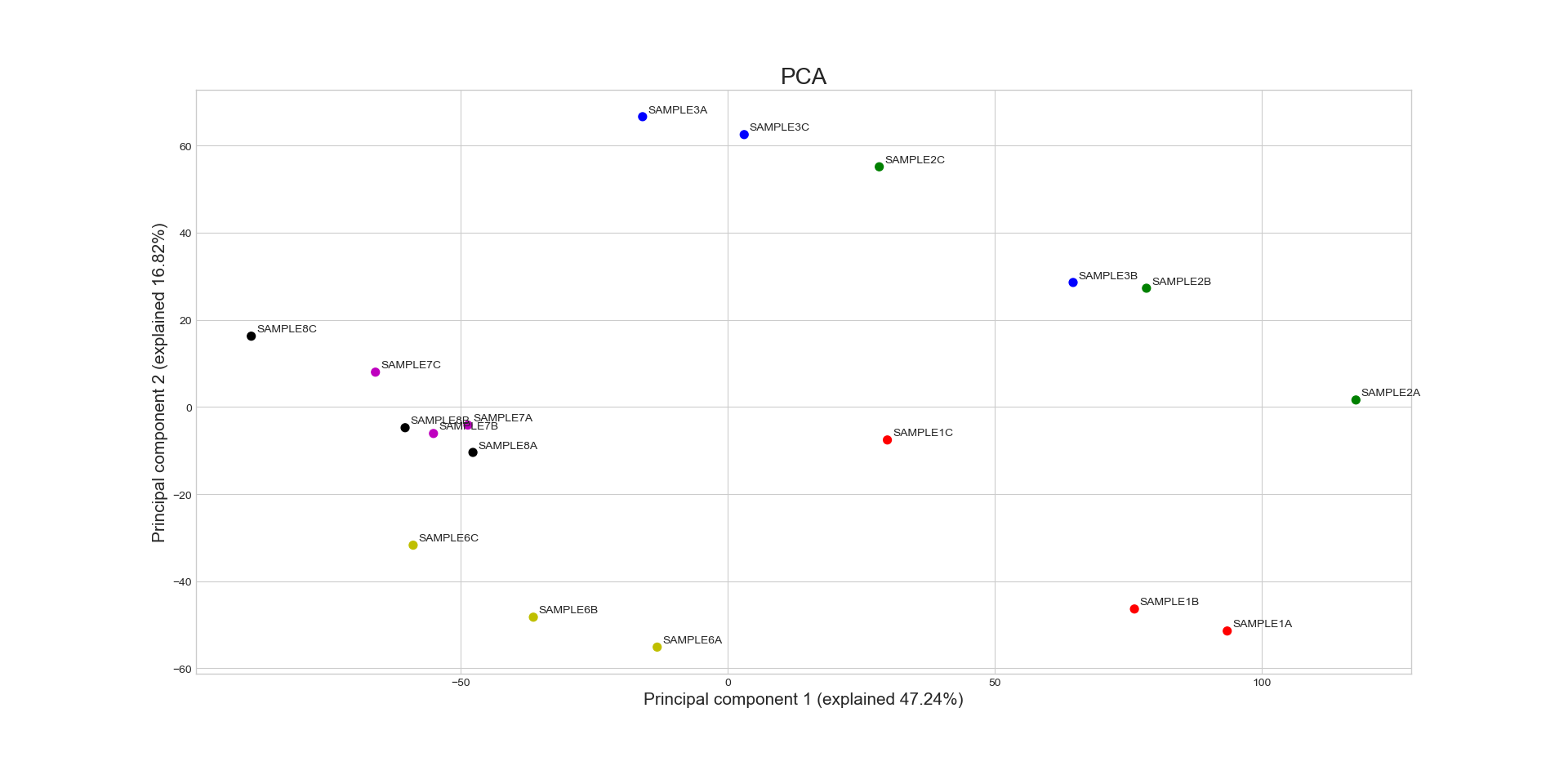
Example plot of pca()